Forecasting Weather (Part 2): Fighting the Data
2025-04-24
Table of Contents
You can read the previous part here.
Overview
Last time was a disaster—I realized I had naively thrown together some code without really knowing what I was doing. We were also missing the key variables, i.e., total precipitation (tp) and total cloud cover (tcc). As I dug into the data, I hit a ton of errors and weird quirks. Things got worse because I was lazy and tried using AI to fix them, but that flopped, so I ditched it halfway through. In this post, I want to walk through all the errors in the order I stumbled into them and finally get to writing some helper functions to plot and visualize the data.
The Right Data
Okay, so having data spaced at 4 hour intervals, for a now-casting project, was far from ideal. So instead I decided to take only 1 month of data but have complete hourly data. I also included total precipitation (tp), total cloud cover (tcc), and geopotential (z) (because we might need it to augment the model with topological data constraints, but we'll see). I also switched to GRIB instead of NetCDF, the reasons are as you will see ahead.
Also we needed the best month to get the data from, with plenty of activity going on, so I picked July 2024.
So the final download request looked like:
import cdsapi
dataset = "reanalysis-era5-single-levels"
request = {
"product_type": ["reanalysis"],
"variable": [
"2m_temperature",
"10m_u_component_of_wind",
"10m_v_component_of_wind",
"relative_humidity",
"total_precipitation",
"surface_pressure",
"total_cloud_cover",
"geopotential"
],
"year": ["2024"],
"month": ["07"],
"day": [
"01", "02", "03", "04", "05", "06", "07", "08", "09", "10",
"11", "12", "13", "14", "15", "16", "17", "18", "19", "20",
"21", "22", "23", "24", "25", "26", "27", "28", "29", "30", "31"
],
"time": [
"00:00", "01:00", "02:00", "03:00", "04:00", "05:00",
"06:00", "07:00", "08:00", "09:00", "10:00", "11:00",
"12:00", "13:00", "14:00", "15:00", "16:00", "17:00",
"18:00", "19:00", "20:00", "21:00", "22:00", "23:00"
],
"data_format": "grib",
"download_format": "unarchived",
"area": "global"
}
client = cdsapi.Client()
client.retrieve(dataset, request).download()
(I saved the file as era5_july2024_global_hourly.grib)
Problem 1: Heterogenous Data
I didn't know the structure of the data I was using, so it was quite the headache to figure stuff out. I did come across this website: ECMWF Parameter Database. But It was already too late.
(NOTE: the next part is before I switched to GRIB)
So excited to try out the new dataset, I did:
import xarray as xr
# Load the NetCDF file
ds = xr.open_dataset("era5_july2024_global_hourly.nc", engine="netcdf4")
# Print the dataset summary
print(ds)
and gracefully got the error:
OSError: NetCDF: Unknown file format
---------------------------------------------------------------------------
KeyError Traceback (most recent call last)
File c:\Users\*****\AppData\Local\Programs\Python\Python312\Lib\site-packages\xarray\backends\file_manager.py:211, in CachingFileManager._acquire_with_cache_info(self, needs_lock)
210 try:
--> 211 file = self._cache[self._key]
212 except KeyError:
File c:\Users\*****\AppData\Local\Programs\Python\Python312\Lib\site-packages\xarray\backends\lru_cache.py:56, in LRUCache.__getitem__(self, key)
55 with self._lock:
---> 56 value = self._cache[key]
57 self._cache.move_to_end(key)
KeyError: [, ('c:\\dev\\weather\\era5_july2024_global_hourly.nc',), 'r', (('clobber', True), ('diskless', False), ('format', 'NETCDF4'), ('persist', False)), 'df0025b7-69bf-4128-8320-5d00cce38840']
During handling of the above exception, another exception occurred:
OSError Traceback (most recent call last)
Cell In[4], line 4
1 import xarray as xr
3 # Load the NetCDF file
----> 4 ds = xr.open_dataset("era5_july2024_global_hourly.nc", engine="netcdf4")
6 # Print the dataset summary
7 print(ds)
File c:\Users\*****\AppData\Local\Programs\Python\Python312\Lib\site-packages\xarray\backends\api.py:687, in open_dataset(filename_or_obj, engine, chunks, cache, decode_cf, mask_and_scale, decode_times, decode_timedelta, use_cftime, concat_characters, decode_coords, drop_variables, inline_array, chunked_array_type, from_array_kwargs, backend_kwargs, **kwargs)
675 decoders = _resolve_decoders_kwargs(
676 decode_cf,
677 open_backend_dataset_parameters=backend.open_dataset_parameters,
(...)
683 decode_coords=decode_coords,
684 )
686 overwrite_encoded_chunks = kwargs.pop("overwrite_encoded_chunks", None)
--> 687 backend_ds = backend.open_dataset(
688 filename_or_obj,
689 drop_variables=drop_variables,
690 **decoders,
691 **kwargs,
692 )
693 ds = _dataset_from_backend_dataset(
694 backend_ds,
695 filename_or_obj,
(...)
705 **kwargs,
706 )
707 return ds
File c:\Users\*****\AppData\Local\Programs\Python\Python312\Lib\site-packages\xarray\backends\netCDF4_.py:666, in NetCDF4BackendEntrypoint.open_dataset(self, filename_or_obj, mask_and_scale, decode_times, concat_characters, decode_coords, drop_variables, use_cftime, decode_timedelta, group, mode, format, clobber, diskless, persist, auto_complex, lock, autoclose)
644 def open_dataset(
645 self,
646 filename_or_obj: str | os.PathLike[Any] | ReadBuffer | AbstractDataStore,
(...)
663 autoclose=False,
664 ) -> Dataset:
665 filename_or_obj = _normalize_path(filename_or_obj)
--> 666 store = NetCDF4DataStore.open(
667 filename_or_obj,
668 mode=mode,
669 format=format,
670 group=group,
671 clobber=clobber,
672 diskless=diskless,
673 persist=persist,
674 auto_complex=auto_complex,
675 lock=lock,
676 autoclose=autoclose,
677 )
679 store_entrypoint = StoreBackendEntrypoint()
680 with close_on_error(store):
File c:\Users\*****\AppData\Local\Programs\Python\Python312\Lib\site-packages\xarray\backends\netCDF4_.py:452, in NetCDF4DataStore.open(cls, filename, mode, format, group, clobber, diskless, persist, auto_complex, lock, lock_maker, autoclose)
448 kwargs["auto_complex"] = auto_complex
449 manager = CachingFileManager(
450 netCDF4.Dataset, filename, mode=mode, kwargs=kwargs
451 )
--> 452 return cls(manager, group=group, mode=mode, lock=lock, autoclose=autoclose)
File c:\Users\*****\AppData\Local\Programs\Python\Python312\Lib\site-packages\xarray\backends\netCDF4_.py:393, in NetCDF4DataStore.__init__(self, manager, group, mode, lock, autoclose)
391 self._group = group
392 self._mode = mode
--> 393 self.format = self.ds.data_model
394 self._filename = self.ds.filepath()
395 self.is_remote = is_remote_uri(self._filename)
File c:\Users\*****\AppData\Local\Programs\Python\Python312\Lib\site-packages\xarray\backends\netCDF4_.py:461, in NetCDF4DataStore.ds(self)
459 @property
460 def ds(self):
--> 461 return self._acquire()
File c:\Users\*****\AppData\Local\Programs\Python\Python312\Lib\site-packages\xarray\backends\netCDF4_.py:455, in NetCDF4DataStore._acquire(self, needs_lock)
454 def _acquire(self, needs_lock=True):
--> 455 with self._manager.acquire_context(needs_lock) as root:
456 ds = _nc4_require_group(root, self._group, self._mode)
457 return ds
File c:\Users\*****\AppData\Local\Programs\Python\Python312\Lib\contextlib.py:137, in _GeneratorContextManager.__enter__(self)
135 del self.args, self.kwds, self.func
136 try:
--> 137 return next(self.gen)
138 except StopIteration:
139 raise RuntimeError("generator didn't yield") from None
File c:\Users\*****\AppData\Local\Programs\Python\Python312\Lib\site-packages\xarray\backends\file_manager.py:199, in CachingFileManager.acquire_context(self, needs_lock)
196 @contextlib.contextmanager
197 def acquire_context(self, needs_lock=True):
198 """Context manager for acquiring a file."""
--> 199 file, cached = self._acquire_with_cache_info(needs_lock)
200 try:
201 yield file
File c:\Users\*****\AppData\Local\Programs\Python\Python312\Lib\site-packages\xarray\backends\file_manager.py:217, in CachingFileManager._acquire_with_cache_info(self, needs_lock)
215 kwargs = kwargs.copy()
216 kwargs["mode"] = self._mode
--> 217 file = self._opener(*self._args, **kwargs)
218 if self._mode == "w":
219 # ensure file doesn't get overridden when opened again
220 self._mode = "a"
File src\\netCDF4\\_netCDF4.pyx:2521, in netCDF4._netCDF4.Dataset.__init__()
File src\\netCDF4\\_netCDF4.pyx:2158, in netCDF4._netCDF4._ensure_nc_success()
**OSError: [Errno -51] NetCDF: Unknown file format: 'c:\\dev\\weather\\era5_july2024_global_hourly.nc'**
I should be crying (I did)—but I thought "maybe the download got corrupted". So I tried about thrice just to make sure— But that wasn't it.
So the thing is— This did not happen with the original test dataset. Which troubled me to no end. I speculated that one of the variables messed up the data, so after several tries selectively removing and downloading and testing, I narrowed the suspect down to Total Precipitation (tp). (This did take a lot of time as the datasets weren't small ~19GB).
Then I noticed that the ERA5 website says that the NetCDF version is "experimental". So I switched over to the GRIB format.
Quite conveniently a library called cfgrib is available, and also works with xarray.
So I tried again with the GRIB version:
import xarray as xr
import numpy as np
import pandas as pd
import os
import matplotlib.pyplot as plt
import cfgrib
grib_ds = xr.open_dataset("era5_july2024_global_hourly.grib", engine="cfgrib")
grib_ds
It didn't work, But more importantly it showed something really insightful:
Skipping variable: paramId==228 shortName='tp'
skipping variable: paramId==228 shortName='tp'
Traceback (most recent call last):
File "c:\Users\*****\anaconda3\envs\era5_env\Lib\site-packages\cfgrib\dataset.py", line 725, in build_dataset_components
dict_merge(variables, coord_vars)
File "c:\Users\*****\anaconda3\envs\era5_env\Lib\site-packages\cfgrib\dataset.py", line 641, in dict_merge
raise DatasetBuildError(
cfgrib.dataset.DatasetBuildError: key present and new value is different: key='time' value=Variable(dimensions=('time',), data=array([1719792000, 1719795600, 1719799200, 1719802800, 1719806400,
1719810000, 1719813600, 1719817200, 1719820800, 1719824400,
1719828000, 1719831600, 1719835200, 1719838800, 1719842400,
1719846000, 1719849600, 1719853200, 1719856800, 1719860400,
1719864000, 1719867600, 1719871200, 1719874800, 1719878400,
1719882000, 1719885600, 1719889200, 1719892800, 1719896400,
1719900000, 1719903600, 1719907200, 1719910800, 1719914400,
1719918000, 1719921600, 1719925200, 1719928800, 1719932400,
1719936000, 1719939600, 1719943200, 1719946800, 1719950400,
1719954000, 1719957600, 1719961200, 1719964800, 1719968400,
1719972000, 1719975600, 1719979200, 1719982800, 1719986400,
1719990000, 1719993600, 1719997200, 1720000800, 1720004400,
1720008000, 1720011600, 1720015200, 1720018800, 1720022400,
1720026000, 1720029600, 1720033200, 1720036800, 1720040400,
1720044000, 1720047600, 1720051200, 1720054800, 1720058400,
1720062000, 1720065600, 1720069200, 1720072800, 1720076400,
1720080000, 1720083600, 1720087200, 1720090800, 1720094400,
1720098000, 1720101600, 1720105200, 1720108800, 1720112400,
1720116000, 1720119600, 1720123200, 1720126800, 1720130400,
1720134000, 1720137600, 1720141200, 1720144800, 1720148400,
1720152000, 1720155600, 1720159200, 1720162800, 1720166400,
1720170000, 1720173600, 1720177200, 1720180800, 1720184400,
1720188000, 1720191600, 1720195200, 1720198800, 1720202400,
1720206000, 1720209600, 1720213200, 1720216800, 1720220400,
1720224000, 1720227600, 1720231200, 1720234800, 1720238400,
1720242000, 1720245600, 1720249200, 1720252800, 1720256400,
1720260000, 1720263600, 1720267200, 1720270800, 1720274400,
1720278000, 1720281600, 1720285200, 1720288800, 1720292400,
1720296000, 1720299600, 1720303200, 1720306800, 1720310400,
1720314000, 1720317600, 1720321200, 1720324800, 1720328400,
1720332000, 1720335600, 1720339200, 1720342800, 1720346400,
1720350000, 1720353600, 1720357200, 1720360800, 1720364400,
1720368000, 1720371600, 1720375200, 1720378800, 1720382400,
1720386000, 1720389600, 1720393200, 1720396800, 1720400400,
1720404000, 1720407600, 1720411200, 1720414800, 1720418400,
1720422000, 1720425600, 1720429200, 1720432800, 1720436400,
1720440000, 1720443600, 1720447200, 1720450800, 1720454400,
1720458000, 1720461600, 1720465200, 1720468800, 1720472400,
1720476000, 1720479600, 1720483200, 1720486800, 1720490400,
1720494000, 1720497600, 1720501200, 1720504800, 1720508400,
1720512000, 1720515600, 1720519200, 1720522800, 1720526400,
1720530000, 1720533600, 1720537200, 1720540800, 1720544400,
1720548000, 1720551600, 1720555200, 1720558800, 1720562400,
1720566000, 1720569600, 1720573200, 1720576800, 1720580400,
1720584000, 1720587600, 1720591200, 1720594800, 1720598400,
1720602000, 1720605600, 1720609200, 1720612800, 1720616400,
1720620000, 1720623600, 1720627200, 1720630800, 1720634400,
1720638000, 1720641600, 1720645200, 1720648800, 1720652400,
1720656000, 1720659600, 1720663200, 1720666800, 1720670400,
1720674000, 1720677600, 1720681200, 1720684800, 1720688400,
1720692000, 1720695600, 1720699200, 1720702800, 1720706400,
1720710000, 1720713600, 1720717200, 1720720800, 1720724400,
1720728000, 1720731600, 1720735200, 1720738800, 1720742400,
1720746000, 1720749600, 1720753200, 1720756800, 1720760400,
1720764000, 1720767600, 1720771200, 1720774800, 1720778400,
1720782000, 1720785600, 1720789200, 1720792800, 1720796400,
1720800000, 1720803600, 1720807200, 1720810800, 1720814400,
1720818000, 1720821600, 1720825200, 1720828800, 1720832400,
1720836000, 1720839600, 1720843200, 1720846800, 1720850400,
1720854000, 1720857600, 1720861200, 1720864800, 1720868400,
1720872000, 1720875600, 1720879200, 1720882800, 1720886400,
1720890000, 1720893600, 1720897200, 1720900800, 1720904400,
1720908000, 1720911600, 1720915200, 1720918800, 1720922400,
1720926000, 1720929600, 1720933200, 1720936800, 1720940400,
1720944000, 1720947600, 1720951200, 1720954800, 1720958400,
1720962000, 1720965600, 1720969200, 1720972800, 1720976400,
1720980000, 1720983600, 1720987200, 1720990800, 1720994400,
1720998000, 1721001600, 1721005200, 1721008800, 1721012400,
1721016000, 1721019600, 1721023200, 1721026800, 1721030400,
1721034000, 1721037600, 1721041200, 1721044800, 1721048400,
1721052000, 1721055600, 1721059200, 1721062800, 1721066400,
1721070000, 1721073600, 1721077200, 1721080800, 1721084400,
1721088000, 1721091600, 1721095200, 1721098800, 1721102400,
1721106000, 1721109600, 1721113200, 1721116800, 1721120400,
1721124000, 1721127600, 1721131200, 1721134800, 1721138400,
1721142000, 1721145600, 1721149200, 1721152800, 1721156400,
1721160000, 1721163600, 1721167200, 1721170800, 1721174400,
1721178000, 1721181600, 1721185200, 1721188800, 1721192400,
1721196000, 1721199600, 1721203200, 1721206800, 1721210400,
1721214000, 1721217600, 1721221200, 1721224800, 1721228400,
1721232000, 1721235600, 1721239200, 1721242800, 1721246400,
1721250000, 1721253600, 1721257200, 1721260800, 1721264400,
1721268000, 1721271600, 1721275200, 1721278800, 1721282400,
1721286000, 1721289600, 1721293200, 1721296800, 1721300400,
1721304000, 1721307600, 1721311200, 1721314800, 1721318400,
1721322000, 1721325600, 1721329200, 1721332800, 1721336400,
1721340000, 1721343600, 1721347200, 1721350800, 1721354400,
1721358000, 1721361600, 1721365200, 1721368800, 1721372400,
1721376000, 1721379600, 1721383200, 1721386800, 1721390400,
1721394000, 1721397600, 1721401200, 1721404800, 1721408400,
1721412000, 1721415600, 1721419200, 1721422800, 1721426400,
1721430000, 1721433600, 1721437200, 1721440800, 1721444400,
1721448000, 1721451600, 1721455200, 1721458800, 1721462400,
1721466000, 1721469600, 1721473200, 1721476800, 1721480400,
1721484000, 1721487600, 1721491200, 1721494800, 1721498400,
1721502000, 1721505600, 1721509200, 1721512800, 1721516400,
1721520000, 1721523600, 1721527200, 1721530800, 1721534400,
1721538000, 1721541600, 1721545200, 1721548800, 1721552400,
1721556000, 1721559600, 1721563200, 1721566800, 1721570400,
1721574000, 1721577600, 1721581200, 1721584800, 1721588400,
1721592000, 1721595600, 1721599200, 1721602800, 1721606400,
1721610000, 1721613600, 1721617200, 1721620800, 1721624400,
1721628000, 1721631600, 1721635200, 1721638800, 1721642400,
1721646000, 1721649600, 1721653200, 1721656800, 1721660400,
1721664000, 1721667600, 1721671200, 1721674800, 1721678400,
1721682000, 1721685600, 1721689200, 1721692800, 1721696400,
1721700000, 1721703600, 1721707200, 1721710800, 1721714400,
1721718000, 1721721600, 1721725200, 1721728800, 1721732400,
1721736000, 1721739600, 1721743200, 1721746800, 1721750400,
1721754000, 1721757600, 1721761200, 1721764800, 1721768400,
1721772000, 1721775600, 1721779200, 1721782800, 1721786400,
1721790000, 1721793600, 1721797200, 1721800800, 1721804400,
1721808000, 1721811600, 1721815200, 1721818800, 1721822400,
1721826000, 1721829600, 1721833200, 1721836800, 1721840400,
1721844000, 1721847600, 1721851200, 1721854800, 1721858400,
1721862000, 1721865600, 1721869200, 1721872800, 1721876400,
1721880000, 1721883600, 1721887200, 1721890800, 1721894400,
1721898000, 1721901600, 1721905200, 1721908800, 1721912400,
1721916000, 1721919600, 1721923200, 1721926800, 1721930400,
1721934000, 1721937600, 1721941200, 1721944800, 1721948400,
1721952000, 1721955600, 1721959200, 1721962800, 1721966400,
1721970000, 1721973600, 1721977200, 1721980800, 1721984400,
1721988000, 1721991600, 1721995200, 1721998800, 1722002400,
1722006000, 1722009600, 1722013200, 1722016800, 1722020400,
1722024000, 1722027600, 1722031200, 1722034800, 1722038400,
1722042000, 1722045600, 1722049200, 1722052800, 1722056400,
1722060000, 1722063600, 1722067200, 1722070800, 1722074400,
1722078000, 1722081600, 1722085200, 1722088800, 1722092400,
1722096000, 1722099600, 1722103200, 1722106800, 1722110400,
1722114000, 1722117600, 1722121200, 1722124800, 1722128400,
1722132000, 1722135600, 1722139200, 1722142800, 1722146400,
1722150000, 1722153600, 1722157200, 1722160800, 1722164400,
1722168000, 1722171600, 1722175200, 1722178800, 1722182400,
1722186000, 1722189600, 1722193200, 1722196800, 1722200400,
1722204000, 1722207600, 1722211200, 1722214800, 1722218400,
1722222000, 1722225600, 1722229200, 1722232800, 1722236400,
1722240000, 1722243600, 1722247200, 1722250800, 1722254400,
1722258000, 1722261600, 1722265200, 1722268800, 1722272400,
1722276000, 1722279600, 1722283200, 1722286800, 1722290400,
1722294000, 1722297600, 1722301200, 1722304800, 1722308400,
1722312000, 1722315600, 1722319200, 1722322800, 1722326400,
1722330000, 1722333600, 1722337200, 1722340800, 1722344400,
1722348000, 1722351600, 1722355200, 1722358800, 1722362400,
1722366000, 1722369600, 1722373200, 1722376800, 1722380400,
1722384000, 1722387600, 1722391200, 1722394800, 1722398400,
1722402000, 1722405600, 1722409200, 1722412800, 1722416400,
1722420000, 1722423600, 1722427200, 1722430800, 1722434400,
1722438000, 1722441600, 1722445200, 1722448800, 1722452400,
1722456000, 1722459600, 1722463200, 1722466800])) new_value=Variable(dimensions=('time',), data=array([1719770400, 1719813600, 1719856800, 1719900000, 1719943200,
1719986400, 1720029600, 1720072800, 1720116000, 1720159200,
1720202400, 1720245600, 1720288800, 1720332000, 1720375200,
1720418400, 1720461600, 1720504800, 1720548000, 1720591200,
1720634400, 1720677600, 1720720800, 1720764000, 1720807200,
1720850400, 1720893600, 1720936800, 1720980000, 1721023200,
1721066400, 1721109600, 1721152800, 1721196000, 1721239200,
1721282400, 1721325600, 1721368800, 1721412000, 1721455200,
1721498400, 1721541600, 1721584800, 1721628000, 1721671200,
1721714400, 1721757600, 1721800800, 1721844000, 1721887200,
1721930400, 1721973600, 1722016800, 1722060000, 1722103200,
1722146400, 1722189600, 1722232800, 1722276000, 1722319200,
1722362400, 1722405600, 1722448800]))
That was it! it was tp. Furthermore, it showed me exactly why the data was different. It is because the timestamps are different. tp was different from the other variables, which made the dataset heterogenous and xarray was unable to load it. (xarray always assumes homogenous data, tried to merge it, and failed).
So what do we do? After some searching I realized that cfgrib has its own load function that can auto-detect the different datasets present in the file. Something like this:
import xarray as xr
import numpy as np
import pandas as pd
import os
import matplotlib.pyplot as plt
import cfgrib
dataset = cfgrib.open_datasets("era5_july2024_global_hourly.grib")
ds1 = dataset[0]
ds2 = dataset[1]
No Errors! Finally. There were in-fact two datasets in that file.
Here is ds1 (everything except tp):
[Sorry I'm lazy but please view these two outputs in lightmode (toggle theme)]
Dataset 1 (Xarray Dataset)
<xarray.Dataset> Size: 19GB Dimensions: (time: 744, latitude: 721, longitude: 1440) Coordinates: number int64 8B 0 * time (time) datetime64[ns] 6kB 2024-07-01 ... 2024-07-31T23:00:00 step timedelta64[ns] 8B 00:00:00 surface float64 8B 0.0 * latitude (latitude) float64 6kB 90.0 89.75 89.5 ... -89.5 -89.75 -90.0 * longitude (longitude) float64 12kB 0.0 0.25 0.5 0.75 ... 359.2 359.5 359.8 valid_time (time) datetime64[ns] 6kB 2024-07-01 ... 2024-07-31T23:00:00 Data variables: z (time, latitude, longitude) float32 3GB ... sp (time, latitude, longitude) float32 3GB ... tcc (time, latitude, longitude) float32 3GB ... u10 (time, latitude, longitude) float32 3GB ... v10 (time, latitude, longitude) float32 3GB ... t2m (time, latitude, longitude) float32 3GB ... Attributes: GRIB_edition: 1 GRIB_centre: ecmf GRIB_centreDescription: European Centre for Medium-Range Weather Forecasts GRIB_subCentre: 0 Conventions: CF-1.7 institution: European Centre for Medium-Range Weather Forecasts
And here is ds2 (only for tp):
Dataset 2 (Xarray Dataset)
<xarray.Dataset> Size: 3GB Dimensions: (time: 63, step: 12, latitude: 721, longitude: 1440) Coordinates: number int64 8B 0 * time (time) datetime64[ns] 504B 2024-06-30T18:00:00 ... 2024-07-31... * step (step) timedelta64[ns] 96B 01:00:00 02:00:00 ... 12:00:00 surface float64 8B 0.0 * latitude (latitude) float64 6kB 90.0 89.75 89.5 ... -89.5 -89.75 -90.0 * longitude (longitude) float64 12kB 0.0 0.25 0.5 0.75 ... 359.2 359.5 359.8 valid_time (time, step) datetime64[ns] 6kB ... Data variables: tp (time, step, latitude, longitude) float32 3GB ... Attributes: GRIB_edition: 1 GRIB_centre: ecmf GRIB_centreDescription: European Centre for Medium-Range Weather Forecasts GRIB_subCentre: 0 Conventions: CF-1.7 institution: European Centre for Medium-Range Weather Forecasts
Writing the plotting helper functions
A wise man once said, "I love looking at graphs and heatmaps". So let's actually, properly, plot our data (unlike last time).
Let's start with total precipitation (tp) as it was giving us the most trouble earlier.
Writing plot_tp
It should be pretty obvious that we're gonna have to do it heatmap/grid style. So what DO we need?, first we might need the data of course, but do we have it in only 2 dimensions? (i.e latitude, longitude, value). Also what are the units. For this let's take a look at the convinient xarray message:
Dataset 2 (Xarray Dataset)
<xarray.Dataset> Size: 3GB Dimensions: (time: 63, step: 12, latitude: 721, longitude: 1440) Coordinates: number int64 8B 0 * time (time) datetime64[ns] 504B 2024-06-30T18:00:00 ... 2024-07-31... * step (step) timedelta64[ns] 96B 01:00:00 02:00:00 ... 12:00:00 surface float64 8B 0.0 * latitude (latitude) float64 6kB 90.0 89.75 89.5 ... -89.5 -89.75 -90.0 * longitude (longitude) float64 12kB 0.0 0.25 0.5 0.75 ... 359.2 359.5 359.8 valid_time (time, step) datetime64[ns] 6kB ... Data variables: tp (time, step, latitude, longitude) float32 3GB ... Attributes: GRIB_edition: 1 GRIB_centre: ecmf GRIB_centreDescription: European Centre for Medium-Range Weather Forecasts GRIB_subCentre: 0 Conventions: CF-1.7 institution: European Centre for Medium-Range Weather Forecasts
We obvserve: 4 dimensions?
Apparently the "time" is divided into 12 steps, and the timestamps, are only given 6 hourly. So do we sum up all the values and only use the 6 hour timestamps? No. that won't be nescessary.
Guess what this outputs:
ds2.tp.time
It is:
xarray.DataArray 'time' (time: 63)
<xarray.DataArray 'time' (time: 63)> Size: 504B array(['2024-06-30T18:00:00.000000000', '2024-07-01T06:00:00.000000000', '2024-07-01T18:00:00.000000000', '2024-07-02T06:00:00.000000000', '2024-07-02T18:00:00.000000000', '2024-07-03T06:00:00.000000000', '2024-07-03T18:00:00.000000000', '2024-07-04T06:00:00.000000000', '2024-07-04T18:00:00.000000000', '2024-07-05T06:00:00.000000000', '2024-07-05T18:00:00.000000000', '2024-07-06T06:00:00.000000000', '2024-07-06T18:00:00.000000000', '2024-07-07T06:00:00.000000000', '2024-07-07T18:00:00.000000000', '2024-07-08T06:00:00.000000000', '2024-07-08T18:00:00.000000000', '2024-07-09T06:00:00.000000000', '2024-07-09T18:00:00.000000000', '2024-07-10T06:00:00.000000000', '2024-07-10T18:00:00.000000000', '2024-07-11T06:00:00.000000000', '2024-07-11T18:00:00.000000000', '2024-07-12T06:00:00.000000000', '2024-07-12T18:00:00.000000000', '2024-07-13T06:00:00.000000000', '2024-07-13T18:00:00.000000000', '2024-07-14T06:00:00.000000000', '2024-07-14T18:00:00.000000000', '2024-07-15T06:00:00.000000000', '2024-07-15T18:00:00.000000000', '2024-07-16T06:00:00.000000000', '2024-07-16T18:00:00.000000000', '2024-07-17T06:00:00.000000000', '2024-07-17T18:00:00.000000000', '2024-07-18T06:00:00.000000000', '2024-07-18T18:00:00.000000000', '2024-07-19T06:00:00.000000000', '2024-07-19T18:00:00.000000000', '2024-07-20T06:00:00.000000000', '2024-07-20T18:00:00.000000000', '2024-07-21T06:00:00.000000000', '2024-07-21T18:00:00.000000000', '2024-07-22T06:00:00.000000000', '2024-07-22T18:00:00.000000000', '2024-07-23T06:00:00.000000000', '2024-07-23T18:00:00.000000000', '2024-07-24T06:00:00.000000000', '2024-07-24T18:00:00.000000000', '2024-07-25T06:00:00.000000000', '2024-07-25T18:00:00.000000000', '2024-07-26T06:00:00.000000000', '2024-07-26T18:00:00.000000000', '2024-07-27T06:00:00.000000000', '2024-07-27T18:00:00.000000000', '2024-07-28T06:00:00.000000000', '2024-07-28T18:00:00.000000000', '2024-07-29T06:00:00.000000000', '2024-07-29T18:00:00.000000000', '2024-07-30T06:00:00.000000000', '2024-07-30T18:00:00.000000000', '2024-07-31T06:00:00.000000000', '2024-07-31T18:00:00.000000000'], dtype='datetime64[ns]') Coordinates: number int64 8B 0 * time (time) datetime64[ns] 504B 2024-06-30T18:00:00 ... 2024-07-31T18... surface float64 8B 0.0 Attributes: long_name: initial time of forecast standard_name: forecast_reference_time
Eh? That didn't say much— how about:
ds2.tp.isel(time=8).values.shape
Output:
(12, 721, 1440)
Interesting! So all the steps can be indexed into after doing the isel.
like:
ds2.tp.isel(time=8).values[0]
Sloppy but works. Also the website says that the precipitation value is in metres (m). Anyways let's get to plotting!
First I made helpers/plotter.py to act as a helper script so we can use the functions in any notebook.
Let's define plot_tp:
def plot_tp(
data: xr.Dataset,
time,
lat_range: tuple = None,
lon_range: tuple = None,
cmap: str = 'viridis',
step: int = None
) -> plt.Figure:
"""
Plot total precipitation (tp) at a specified time.
Parameters
----------
data : xarray.Dataset
Must contain variable 'tp' with dims (time, step, latitude, longitude).
time : str or numpy.datetime64
Timestamp to select (nearest match).
lat_range : tuple (min_lat, max_lat), optional
Latitude subsetting range.
lon_range : tuple (min_lon, max_lon), optional
Longitude subsetting range.
cmap : str or Colormap, default 'viridis'
Colormap for precipitation shading.
Returns
-------
plt.Figure
The Matplotlib Figure object of the precipitation plot.
"""
Then we just select the time from the array:
da = data.tp.sel(time=time, method='nearest')
arr = da[0] if step else (da.fillna(0).sum(dim='step') * 1000).compute()
Why the .compute()? I'll explain later, it was mainly for debugging but I decided to leave it in there.
Also the next part was done over a lot of trial and error, and in retrospect has grown way too complex, but it works, with edge cases too so I don't want to break it.
if lat_range:
arr = arr.sel(latitude=_slice_coord(data.latitude.values, *lat_range))
if lon_range:
arr = arr.sel(longitude=_slice_coord(data.longitude.values, *lon_range))
if arr.size == 0:
msg = (
f"Empty selection: lat {data.latitude.min().item()}–{data.latitude.max().item()}, "
f"lon {data.longitude.min().item()}–{data.longitude.max().item()}"
)
raise ValueError(msg)
if np.all(np.isnan(arr)):
raise ValueError("All values are NaN for this time/region.")
vmin, vmax = arr.min().item(), arr.max().item()
norm = plt.Normalize(vmin=vmin, vmax=vmax) if vmax > vmin else None
Pretty straight forward error handling (Maybe not, it took me a few days to get right so—). cleaning NaNs, No empty Selections etc.
What is _slice_coord()? It's to deal with wrap arounds (No idea why I added this, I'm the only one who is going to use it anyway). Here is the code:
def _slice_coord(
coord: np.ndarray,
min_val: float,
max_val: float
) -> slice:
"""
Compute a robust slice for an array of coordinate values that may be
in ascending or descending order.
Parameters
----------
coord : 1D numpy array
Coordinate values (e.g., latitude or longitude).
min_val : float
Desired minimum coordinate.
max_val : float
Desired maximum coordinate.
Returns
-------
slice
Slice object for selecting the desired range from an xarray DataArray.
"""
ascending = coord[0] < coord[-1]
if not ascending:
rev = coord[::-1]
i_min = len(coord) - 1 - np.searchsorted(rev, min_val, 'left')
i_max = len(coord) - 1 - np.searchsorted(rev, max_val, 'right')
start, stop = sorted((coord[i_min], coord[i_max]), reverse=True)
else:
i_min = np.searchsorted(coord, min_val, 'left')
i_max = np.searchsorted(coord, max_val, 'right') - 1
start, stop = sorted((coord[i_min], coord[i_max]))
return slice(start, stop)
Now the plotting part: (We're back in plot_tp())
lons2d, lats2d = np.meshgrid(arr.longitude, arr.latitude)
fig = plt.figure(figsize=(12, 6))
ax = fig.add_subplot(1, 1, 1, projection=ccrs.PlateCarree())
ax.coastlines(resolution='50m', color='gray')
ax.add_feature(cfeature.BORDERS, linestyle=':', linewidth=0.5)
pcm = ax.pcolormesh(
lons2d, lats2d, arr,
transform=ccrs.PlateCarree(), cmap=cmap,
norm=norm, shading='auto'
)
cbar = fig.colorbar(pcm, ax=ax, orientation='vertical', pad=0.02)
cbar.set_label('Total Precipitation (mm)')
time_actual = np.datetime_as_string(arr.time.values, unit='h')
ax.set_title(f'Total Precipitation for {time_actual}')
if lat_range or lon_range:
extent = [arr.longitude.min(), arr.longitude.max(), arr.latitude.min(), arr.latitude.max()]
ax.set_extent(extent, crs=ccrs.PlateCarree())
plt.tight_layout()
plt.show()
return fig
Most of this is pretty standard procedure— Again I added the normalization and stuff but I think it was just fine without.
So finally our complicated function looks something like:
def plot_tp(
data: xr.Dataset,
time,
lat_range: tuple = None,
lon_range: tuple = None,
cmap: str = 'viridis',
step: int = None
) -> plt.Figure:
"""
Plot total precipitation (tp) at a specified time.
Parameters
----------
data : xarray.Dataset
Must contain variable 'tp' with dims (time, step, latitude, longitude).
time : str or numpy.datetime64
Timestamp to select (nearest match).
lat_range : tuple (min_lat, max_lat), optional
Latitude subsetting range.
lon_range : tuple (min_lon, max_lon), optional
Longitude subsetting range.
cmap : str or Colormap, default 'viridis'
Colormap for precipitation shading.
Returns
-------
plt.Figure
The Matplotlib Figure object of the precipitation plot.
"""
da = data.tp.sel(time=time, method='nearest')
arr = da[0] if step else (da.fillna(0).sum(dim='step') * 1000).compute()
if lat_range:
arr = arr.sel(latitude=_slice_coord(data.latitude.values, *lat_range))
if lon_range:
arr = arr.sel(longitude=_slice_coord(data.longitude.values, *lon_range))
if arr.size == 0:
msg = (
f"Empty selection: lat {data.latitude.min().item()}–{data.latitude.max().item()}, "
f"lon {data.longitude.min().item()}–{data.longitude.max().item()}"
)
raise ValueError(msg)
if np.all(np.isnan(arr)):
raise ValueError("All values are NaN for this time/region.")
vmin, vmax = arr.min().item(), arr.max().item()
norm = plt.Normalize(vmin=vmin, vmax=vmax) if vmax > vmin else None
lons2d, lats2d = np.meshgrid(arr.longitude, arr.latitude)
fig = plt.figure(figsize=(12, 6))
ax = fig.add_subplot(1, 1, 1, projection=ccrs.PlateCarree())
ax.coastlines(resolution='50m', color='gray')
ax.add_feature(cfeature.BORDERS, linestyle=':', linewidth=0.5)
pcm = ax.pcolormesh(
lons2d, lats2d, arr,
transform=ccrs.PlateCarree(), cmap=cmap,
norm=norm, shading='auto'
)
cbar = fig.colorbar(pcm, ax=ax, orientation='vertical', pad=0.02)
cbar.set_label('Total Precipitation (mm)')
time_actual = np.datetime_as_string(arr.time.values, unit='h')
ax.set_title(f'Total Precipitation for {time_actual}')
if lat_range or lon_range:
extent = [arr.longitude.min(), arr.longitude.max(), arr.latitude.min(), arr.latitude.max()]
ax.set_extent(extent, crs=ccrs.PlateCarree())
plt.tight_layout()
plt.show()
return fig
And finally some plots:
from helpers.plotter import *
plot_tp(ds2, "2024-07-02T18:00:00", lat_range=(5.0, 40.0), lon_range=(60, 100), step=0)
plot_tp(ds2, "2024-07-02T18:00:00")
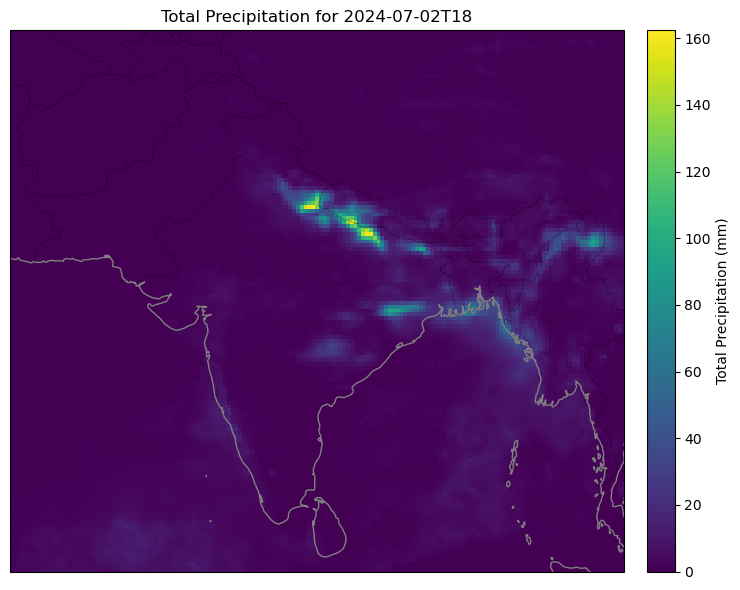
Yeah the global one isn't the best.
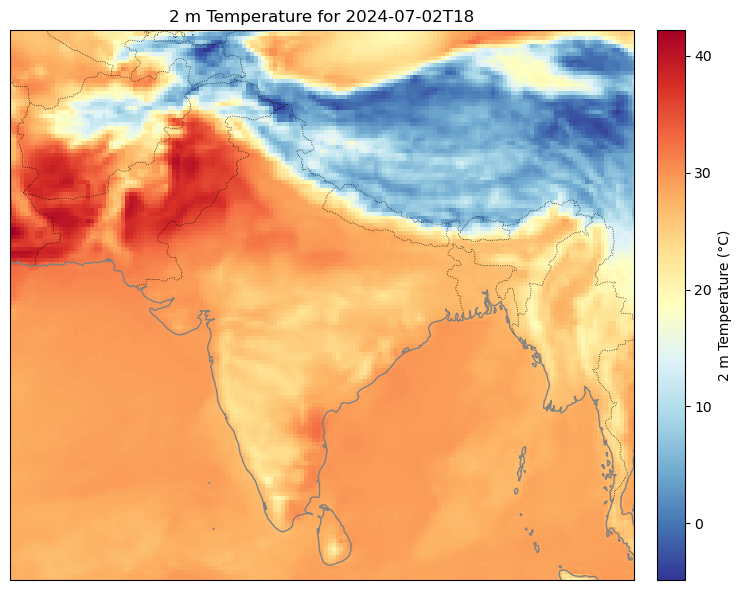
Anyways we can do the same for t2m and tcc, just removing the step functionality.
Here are some plots I got:
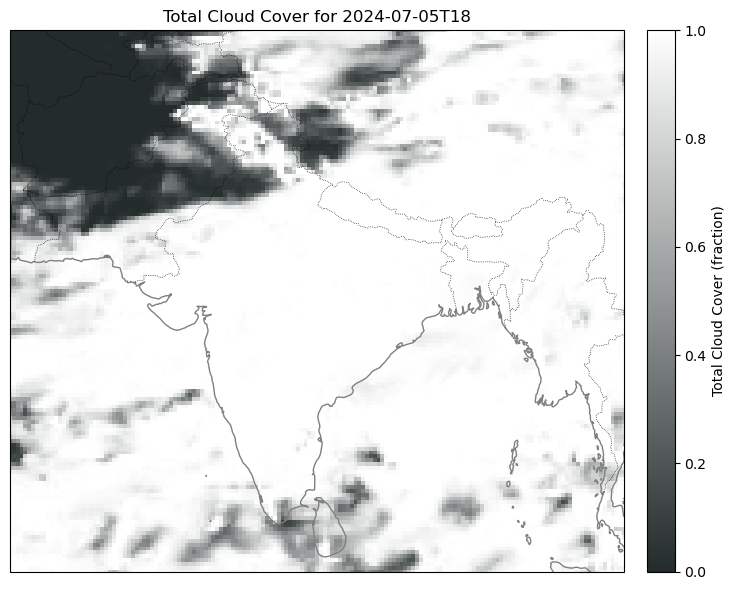
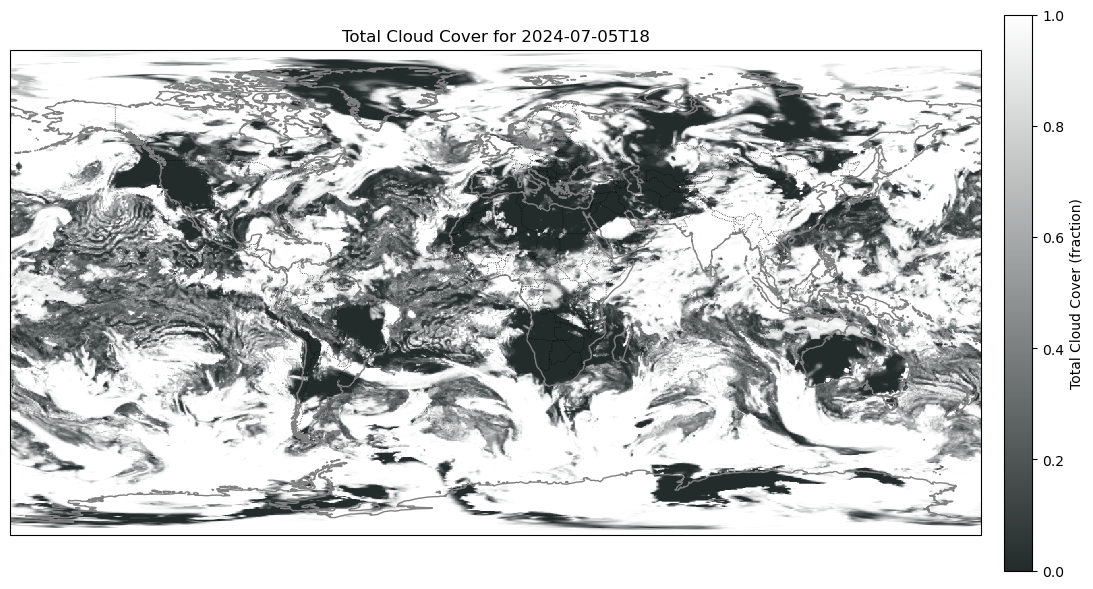
Problem 2: Too Big GRIB No Work.
[I still haven't tackled this problem, but it's more of just chunking the dataset, and I'm gonna figure that out later while dataloading anyways]
You might've noticed that we never indexed above 2nd July 2024, even though we downloaded the entire month's data? Let's try that, say we take 15th July 2024, with something like:
from helpers.plotter import *
plot_tp(ds2, "2024-07-15T18:00:00", lat_range=(5.0, 40.0), lon_range=(60, 100), step=0)
we get this error:
OSError: [Errno 22] Invalid argument
---------------------------------------------------------------------------
OSError Traceback (most recent call last)
Cell In[7], line 3
1 from helpers.plotter import *
----> 3 plot_tp(ds2, "2024-07-15T18:00:00", lat_range=(5.0, 40.0), lon_range=(60, 100), step=0)
File c:\dev\weather\helpers\plotter.py:136, in plot_tp(data, time, lat_range, lon_range, cmap, step)
114 """
115 Plot total precipitation (tp) at a specified time.
116
(...) 133 The Matplotlib Figure object of the precipitation plot.
134 """
135 da = data.tp.sel(time=time, method='nearest')
--> 136 arr = da[0] if step else (da.fillna(0).sum(dim='step') * 1000).compute()
138 if lat_range:
139 arr = arr.sel(latitude=_slice_coord(data.latitude.values, *lat_range))
File c:\Users\*****\anaconda3\envs\era5_env\Lib\site-packages\xarray\core\dataarray.py:3525, in DataArray.fillna(self, value)
3520 if utils.is_dict_like(value):
3521 raise TypeError(
3522 "cannot provide fill value as a dictionary with "
3523 "fillna on a DataArray"
3524 )
-> 3525 out = ops.fillna(self, value)
3526 return out
File c:\Users\*****\anaconda3\envs\era5_env\Lib\site-packages\xarray\core\ops.py:148, in fillna(data, other, join, dataset_join)
124 """Fill missing values in this object with data from the other object.
125 Follows normal broadcasting and alignment rules.
126
(...) 144 - "right": take only variables from the last object
145 """
146 from xarray.core.computation import apply_ufunc
--> 148 return apply_ufunc(
149 duck_array_ops.fillna,
150 data,
151 other,
152 join=join,
153 dask="allowed",
154 dataset_join=dataset_join,
155 dataset_fill_value=np.nan,
156 keep_attrs=True,
157 )
File c:\Users\*****\anaconda3\envs\era5_env\Lib\site-packages\xarray\core\computation.py:1265, in apply_ufunc(func, input_core_dims, output_core_dims, exclude_dims, vectorize, join, dataset_join, dataset_fill_value, keep_attrs, kwargs, dask, output_dtypes, output_sizes, meta, dask_gufunc_kwargs, on_missing_core_dim, *args)
1263 # feed DataArray apply_variable_ufunc through apply_dataarray_vfunc
1264 elif any(isinstance(a, DataArray) for a in args):
-> 1265 return apply_dataarray_vfunc(
1266 variables_vfunc,
1267 *args,
1268 signature=signature,
1269 join=join,
1270 exclude_dims=exclude_dims,
1271 keep_attrs=keep_attrs,
1272 )
1273 # feed Variables directly through apply_variable_ufunc
1274 elif any(isinstance(a, Variable) for a in args):
File c:\Users\*****\anaconda3\envs\era5_env\Lib\site-packages\xarray\core\computation.py:307, in apply_dataarray_vfunc(func, signature, join, exclude_dims, keep_attrs, *args)
302 result_coords, result_indexes = build_output_coords_and_indexes(
303 args, signature, exclude_dims, combine_attrs=keep_attrs
304 )
306 data_vars = [getattr(a, "variable", a) for a in args]
--> 307 result_var = func(*data_vars)
309 out: tuple[DataArray, ...] | DataArray
310 if signature.num_outputs > 1:
File c:\Users\*****\anaconda3\envs\era5_env\Lib\site-packages\xarray\core\computation.py:729, in apply_variable_ufunc(func, signature, exclude_dims, dask, output_dtypes, vectorize, keep_attrs, dask_gufunc_kwargs, *args)
722 broadcast_dims = tuple(
723 dim for dim in dim_sizes if dim not in signature.all_core_dims
724 )
725 output_dims = [broadcast_dims + out for out in signature.output_core_dims]
727 input_data = [
728 (
--> 729 broadcast_compat_data(arg, broadcast_dims, core_dims)
730 if isinstance(arg, Variable)
731 else arg
732 )
733 for arg, core_dims in zip(args, signature.input_core_dims, strict=True)
734 ]
736 if any(is_chunked_array(array) for array in input_data):
737 if dask == "forbidden":
File c:\Users\*****\anaconda3\envs\era5_env\Lib\site-packages\xarray\core\computation.py:650, in broadcast_compat_data(variable, broadcast_dims, core_dims)
645 def broadcast_compat_data(
646 variable: Variable,
647 broadcast_dims: tuple[Hashable, ...],
648 core_dims: tuple[Hashable, ...],
649 ) -> Any:
--> 650 data = variable.data
652 old_dims = variable.dims
653 new_dims = broadcast_dims + core_dims
File c:\Users\*****\anaconda3\envs\era5_env\Lib\site-packages\xarray\core\variable.py:474, in Variable.data(self)
472 return self._data
473 elif isinstance(self._data, indexing.ExplicitlyIndexed):
--> 474 return self._data.get_duck_array()
475 else:
476 return self.values
File c:\Users\*****\anaconda3\envs\era5_env\Lib\site-packages\xarray\core\indexing.py:840, in MemoryCachedArray.get_duck_array(self)
839 def get_duck_array(self):
--> 840 self._ensure_cached()
841 return self.array.get_duck_array()
File c:\Users\*****\anaconda3\envs\era5_env\Lib\site-packages\xarray\core\indexing.py:837, in MemoryCachedArray._ensure_cached(self)
836 def _ensure_cached(self):
--> 837 self.array = as_indexable(self.array.get_duck_array())
File c:\Users\*****\anaconda3\envs\era5_env\Lib\site-packages\xarray\core\indexing.py:794, in CopyOnWriteArray.get_duck_array(self)
793 def get_duck_array(self):
--> 794 return self.array.get_duck_array()
File c:\Users\*****\anaconda3\envs\era5_env\Lib\site-packages\xarray\core\indexing.py:657, in LazilyIndexedArray.get_duck_array(self)
653 array = apply_indexer(self.array, self.key)
654 else:
655 # If the array is not an ExplicitlyIndexedNDArrayMixin,
656 # it may wrap a BackendArray so use its __getitem__
--> 657 array = self.array[self.key]
659 # self.array[self.key] is now a numpy array when
660 # self.array is a BackendArray subclass
661 # and self.key is BasicIndexer((slice(None, None, None),))
662 # so we need the explicit check for ExplicitlyIndexed
663 if isinstance(array, ExplicitlyIndexed):
File c:\Users\*****\anaconda3\envs\era5_env\Lib\site-packages\cfgrib\xarray_plugin.py:163, in CfGribArrayWrapper.__getitem__(self, key)
159 def __getitem__(
160 self,
161 key: xr.core.indexing.ExplicitIndexer,
162 ) -> np.ndarray:
--> 163 return xr.core.indexing.explicit_indexing_adapter(
164 key, self.shape, xr.core.indexing.IndexingSupport.BASIC, self._getitem
165 )
File c:\Users\*****\anaconda3\envs\era5_env\Lib\site-packages\xarray\core\indexing.py:1018, in explicit_indexing_adapter(key, shape, indexing_support, raw_indexing_method)
996 """Support explicit indexing by delegating to a raw indexing method.
997
998 Outer and/or vectorized indexers are supported by indexing a second time
(...) 1015 Indexing result, in the form of a duck numpy-array.
1016 """
1017 raw_key, numpy_indices = decompose_indexer(key, shape, indexing_support)
-> 1018 result = raw_indexing_method(raw_key.tuple)
1019 if numpy_indices.tuple:
1020 # index the loaded np.ndarray
1021 indexable = NumpyIndexingAdapter(result)
File c:\Users\*****\anaconda3\envs\era5_env\Lib\site-packages\cfgrib\xarray_plugin.py:172, in CfGribArrayWrapper._getitem(self, key)
167 def _getitem(
168 self,
169 key: T.Tuple[T.Any, ...],
170 ) -> np.ndarray:
171 with self.datastore.lock:
--> 172 return self.array[key]
File c:\Users\*****\anaconda3\envs\era5_env\Lib\site-packages\cfgrib\dataset.py:373, in OnDiskArray.__getitem__(self, item)
371 continue
372 # NOTE: fill a single field as found in the message
--> 373 message = self.index.get_field(message_ids[0]) # type: ignore
374 values = get_values_in_order(message, array_field[tuple(array_field_indexes)].shape)
375 array_field.__getitem__(tuple(array_field_indexes)).flat[:] = values
File c:\Users\*****\anaconda3\envs\era5_env\Lib\site-packages\cfgrib\messages.py:488, in FieldsetIndex.get_field(self, message_id)
487 def get_field(self, message_id: T.Any) -> abc.Field:
--> 488 return ComputedKeysAdapter(self.fieldset[message_id], self.computed_keys)
File c:\Users\*****\anaconda3\envs\era5_env\Lib\site-packages\cfgrib\messages.py:345, in FileStream.__getitem__(self, item)
343 def __getitem__(self, item: T.Optional[OffsetType]) -> Message:
344 with open(self.path, "rb") as file:
--> 345 return self.message_from_file(file, offset=item)
File c:\Users\*****\anaconda3\envs\era5_env\Lib\site-packages\cfgrib\messages.py:341, in FileStream.message_from_file(self, file, offset, **kwargs)
339 def message_from_file(self, file, offset=None, **kwargs):
340 # type: (T.IO[bytes], T.Optional[OffsetType], T.Any) -> Message
--> 341 return Message.from_file(file, offset, **kwargs)
File c:\Users\*****\anaconda3\envs\era5_env\Lib\site-packages\cfgrib\messages.py:94, in Message.from_file(cls, file, offset, **kwargs)
92 offset, field_in_message = offset
93 if offset is not None:
---> 94 file.seek(offset)
95 codes_id = None
96 if field_in_message == 0:
**OSError: [Errno 22] Invalid argument**
What? Why? I rechecked, even redownloaded the dataset... so we definitely DO have the data. Then what went wrong?
It has something to do with how cfgrib works (on windows especially)
You see, cfgrib uses eccodes, a C library under the hood. What's the problem with that? Quite a big one actually.
So first of all, we should notice that whenever cfgrib loads our dataset it creates an index file, like era5_july2024_global_hourly.grib.5b7b6.idx. Now a plausible explanation might be that they are still using the long datatype instead of fixed ones. This would actually cause a lot quirky platform problems. On Windows this might mean 32-bit offsets, which roughly translates to <2GB of data. So whenever we seek past 2GB, it overflows, possibly goes negative, hence the error. This is highly speculative though, and my rudimentary C++ skills should not be relied on.
I mentioned .compute() before, this is why it was needed. I was trying to test whether we can actually carry out computations on data beyond 5th july, it would also force xarray to not lazy load it. But either way it didn't work.
Random Rant: Ok so I accidentally deleted the 19GB dataset I downloaded for the 4th time now- This has cost me more time than all the actual coding combined! I was trying to write a demo notebook to show how to download it using the API, but guess what I executed the cell, out of muscle memory, and when I stopped it, I realized it was too late because it had already deleted the file with the same name... True Horror story based on real events. I really need to add some fail-safes, yep let's add that to the agenda next time (Future me: It happened again, this time I fat fingered in the command prompt itself)
Ok back to the topic. So I don't want to bother fixing this right now as we won't be loading data this way anyways, we will be chunking it in the future and trying to feed it to pytorch DataLoader and what not.
Conclusion & Plans
In this post we explored the actual structure of the data, and also touched up on some plotting, solving a few errors along the way.
I have realized that it is imperative that we must first draft up a Project-Spec, so I will be doing that next time. Next-to-Next time, I want to try to set up WandB (Never used it before, but I'm willing to join the cult), and possibly get to DataLoading. Of course, we must also cover the augmentation of data with stuff like solar angle, and topology/geopotential, for physics-informed loss purposes.
You can read the next part here (Not Released Yet)